
French genetics
for cattle, sheep
and goat industries
-
Languages :







- Subscribe to the newsletter RSS
-
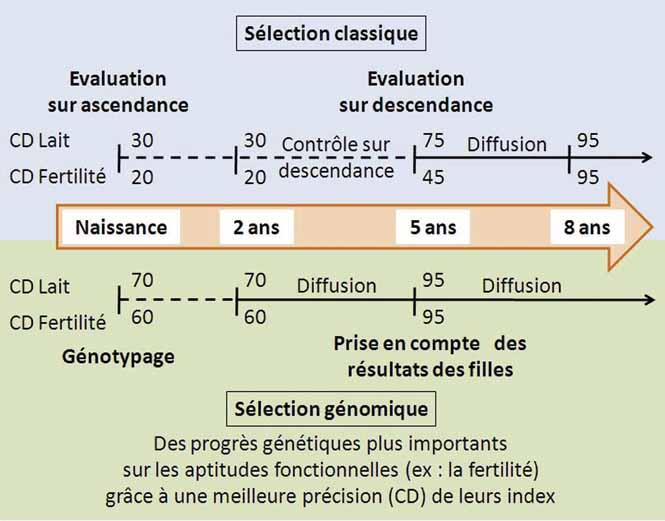
A step-by-step selection process
Genomics

A revolution on the way
The fact that France has successfully integrated genomics science into its selection programmes is a major technological leap forward. The size of the reference breed populations coupled with decade-long optimization of the scientific methods used enables robustly reliable genomics evaluations on every trait conventionally evaluated on progeny.
The value of this genotyping dataset can be further exploited to generate greater genetic progress, broader range of genetic offer and even sharper indexing of the functional traits. Genomics-enhanced evaluation therefore opens up new perspectives, including in terms of selecting for functional traits.
France among the world leaders
France was one of the first countries in the world to gain official international validation from Interbull on its genomics evaluation protocols.
This position at the leading edge of genomic selection has been built up through long-term close cooperation between members of the France Génétique Elevage network. Currently this cooperation brings together the national federation of selection-industry businesses (UNCEIA), the selection businesses themselves, the Institut de l’Elevage [French national Livestock Institute], and the INRA [French national institute for agronomics research] which delivers the basic research component.
From as early as 2001, the breed selection programmes for the Holstein, Montbéliard and Normande have been constantly optimized (candidate bulls screening ahead of on-station testing, etc.) to integrate genomics breakthroughs as soon as they are confirmed.
Since 2008, capitalizing on this policy of continuously integrating technological innovations, the selection programmes have been using use genomics data at every phase of the selection chain: choice of bull parents, males shortlisted for the testing station, selection of the bulls for planned mating...
In June 2009, this next-generation genetics data was unlocked for all breeders via official published release of the first Holstein, Montbéliard and Normande bull genomics indexes.
Every trait traditionally evaluated on progeny now has its own genomics evaluation, from dairy productivity (milk quantity and quality) to animal morphology and on to functional traits (traits liable to cut production costs, such as cow fertility, mastitis resistance, and so on).
A fully-optimized evaluation method
The French genomics-enhanced evaluation method has been comprehensively optimized through ultra-accurate progeny tests and major investment in methodological input. The reliability of these genomic indexes stems from the hugely robust size of the reference populations: 1,250 Normande-breed bulls, 1,500 Montbéliard-breed bulls and 18,300 Holstein-breed bulls, all within the framework of the pan-European EuroGenomics project.
Since 2009, impelled by an UNCEIA-led initiative, Europe’s leading Holstein breed selection businesses joined forces to pool the reference population datasets (the EuroGenomics project).
Counting over 18,000 fully genotyped and progeny-tested purebred Holstein bulls (and 19 millions daughters!), this vast one-of-its kind reference population has been built on the best genetic material that Europe and North America can provide.
Although each country still works with its own evaluation methods, the EuroGenomics project partners now co-use a pooled reference population. Compared to genomics evaluation based on a national-scale reference population the gain in accuracy is 10%.
A broader offer for greater sustainability
Thanks to integration of genomics-based technologies, French genetic selection programmes are now able to offer an extended range of high-performance sires with sharper indexes on functional traits. These sires open up new perspectives for farmers in terms of improved genetic gains on traits that are pivotal to livestock system sustainability.
First, there was the conventional offer of progeny-tested bulls. Now, there is also a new offer of genomically-selected bulls in all three core French dairy breeds.
Following a long period dominated by the sons of just a handful of stud sires, the wide panel of Holstein sires now available offer much broader genetic diversity. Their profiles are adapted to a range of objectives: improved milk production, improved morphology, or improved functional traits.
This range, put together with high-ISU-index (all-round composite index) breed sires, also includes bulls boasting other sought-after criteria (redding, naturally polled…).
The Montbéliard and Normande breeds have also been range-extended and rejuvenated to offer recent-pedigree sires and sires presenting profiles that showcase the hybrid vigour of their breed mix (good all-round value set, lifespan, mastitis resistance).
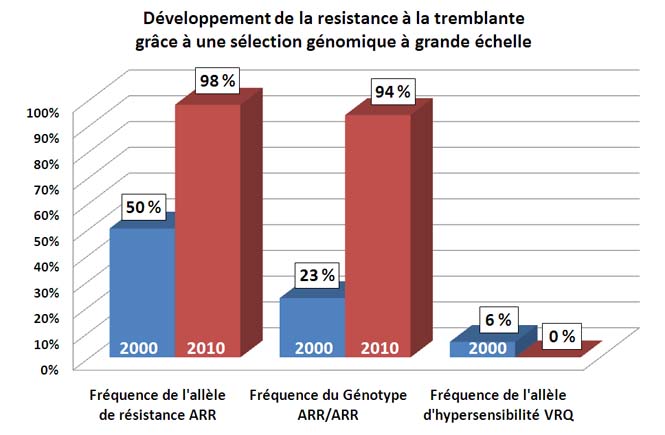
Large-scale selection of a major sheep gene
The National Genetic Improvement Plan for Scrapie-Resistant Sheep, encompassing provides to all French sheep breeds a unique case-illustration of very-large-scale selection for a major gene : PrP gene that confers varying degrees of scrapie resistance.
The scheme coordinated the genotyping of over 670,000 sheep in the space of 6 years, with the onus on eliminating breeder rams carrying the PrP gene susceptibility alleles (VRQ and AHQ) and disseminating those carrying the resistance alleles (ARR).
The results command attention. Since 2008, there is not a single ram from any farm in the sheep meat breed selection population that possesses the VRQ scrapie hypersensitivity allele. Furthermore, over 95% of them are scrapie-resistant (ARR/ARR genotype).
Consolidated assets and new perspectives
The other dairy and beef cattle breeds, as well as sheep and goat breeds, are soon to reap the benefits of the genomics revolution.
With experience built in and new output from large-scale scientific research programmes, genomics-based evaluations are set to be extended to encompass criteria that conventional selection programmes are incapable to integrate, such as milk composition components with positive (or negative) effects on human health (fatty acids, etc.), disease resistance, meat quality (tenderness, marbling, flavour, and more).
The critical prerequisite for developing robust predictive formulae is to work with a reference population comprising fully genotyped animals on which these specific traits have also been measured.
That is the main purpose of 2 programmes led by the INRA, the UNCEIA, the Institut de l’Elevage and selection-industry businesses.
- Phénofinlait (advanced compositional analysis of the milk from 20,000 genotyped cows, goats and sheep)
- Qualvigène (meat quality analysis on over 3,000 young bulls genotyped).
As with the Holstein, Montbéliard and Normande breeds, the next generation of breed sires will not be marketed until their genomics-enhanced evaluations gained proven reliability, international acceptance and recognition.
These are the basic condition to achieve our quality expectation.
Key figures
- 15 years of scientific genomics research
- 10 years of genomic application in selection programmes
- 750 000 animals already genotyped for selection purposes
- 3 breeds benefitting from genomic evaluations
- 15 breeds soon able to benefit from genomic evaluations
- 1 major gene selected for at large-scale in the sheep species
- 2 large-scale programmes targeting product quality
Document(s)
Cart documents to download directly or to add to your cart for download at the end of visit


































